Variational autoencoding of gene landscapes during mouse CNS development uncovers layered roles of Polycomb Repressor Complex 2¶
Information¶
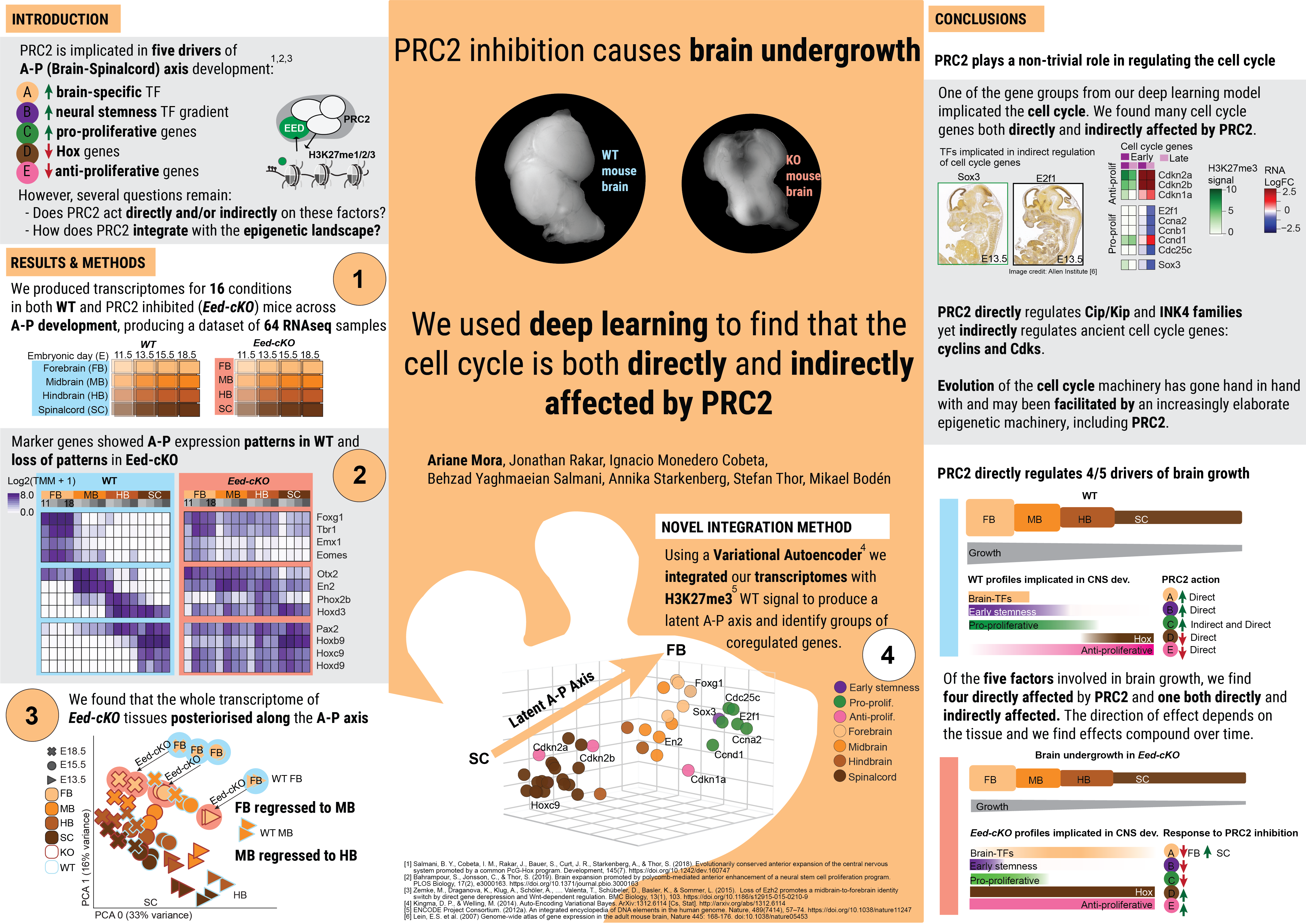
This site hosts the information associated with the paper: Deep Learning Decodes Multi-layered Roles of Polycomb Repressor Complex 2 During Mouse CNS Development. Here we provide the code and data used for all the analyses in the paper and link to the packages we developed as part of producing the paper.
Links to analyses and data¶
Preprint: bioRxiv
HTML outputs are available on the left panel
Code and processed data is available at: mouseCNS_vae
Raw RNA-seq files are available at the NCBI/Gene Expression Omnibus under the accession GSE123331
Links to developed packages and tutorials¶
Reproducible Variational autoencoder: scivae
Epigenetic annotation to genes in python scie2g
Biomart python wrapper scibiomart
Visualisation library for chart formatting sciviso
Updates since review¶
We’ve just been through our first round of revisions (November 2021), and we have updated several things:
We created an interactive website where you can interact with the data.
Made the data easier to download without having to run anything.
Added all marked anterior genes (121) to the analysis where we test for conservation of tissue specificity between mouse and humans
Added a comparison to identify if there are more blood vessel staining under Eed-cKO.
The manuscript also has some nice updates where we put everything in context a bit better and highlighted the novelty :)
Places where this (or a package we developed for this) has been presented¶
Date |
Conference |
Type |
|---|---|---|
28 April 2021 |
Melbourne bioinformatics seminar series |
Presentation |
25 May 2021 |
Poster |
|
25 July 2021 |
Poster |
|
10 November 2021 |
Lightning talk |
|
18 November 2021 |
Poster |
|
19 November 2021 |
SCMB School of Chemistry and Molecular Biosciences symposium |
Presentation |
22 November 2021 |
ABACBS Australian Bioinformatics And Computational Biology Society |
Lightning talk |
Authors¶
Ariane Mora1, Jonathan Rakar3, Ignacio Monedero Cobeta3*, Behzad Yaghmaeian Salmani3#, Annika Starkenberg3, Stefan Thor2,3$, Mikael Bodén1
1) School of Chemistry and Molecular Biosciences 2), and School of Biomedical Sciences, University of Queensland, St Lucia QLD 4072, Australia. 3) Department of Clinical and Experimental Medicine, Linkoping University, SE-58185, Linkoping, Sweden.
Current addresses: #) Department of Cell and Molecular Biology, Karolinska Institute, SE-171 65, Stockholm, Sweden. *Department of Physiology, Universidad Autonoma de Madrid, Madrid, Spain. $) School of Biomedical Sciences, University of Queensland, St Lucia QLD 4072, Australia
Abstract¶
A prominent aspect of most, if not all, central nervous systems (CNSs) is that anterior regions (brain) are larger than posterior ones (spinal cord). Studies in Drosophila and mouse have revealed that the Polycomb Repressor Complex 2 (PRC2) acts by several mechanisms to promote anterior CNS expansion. However, it is unclear if PRC2 acts directly and/or indirectly upon key downstream genes, what the full spectrum of PRC2 action is during embryonic CNS development and how PRC2 integrates with the epigenetic landscape. We removed PRC2 function from the developing mouse CNS, by mutating the key gene Eed, and generated spatio-temporal transcriptomic data. We developed a bioinformatics workflow that incorporates standard statistical analyses with machine learning to integrate the transcriptomic response to PRC2 inactivation with epigenetic information from ENCODE. This multi-variate analysis corroborates the central involvement of PRC2 in anterior CNS expansion, and reveals layered regulation via PRC2. These findings uncover a differential logic for the role of PRC2 upon functionally distinct gene categories that drive CNS anterior expansion. To support the analysis of emerging multi-modal datasets, we provide a novel bioinformatics package that can disentangle regulatory underpinnings of heterogeneous biological processes. Code for our scivae package is available at sci-vae.
Getting in touch¶
Please contact ST (s.thor@uq.edu.au), MB (m.boden@uq.edu.au), and AM (a.mora@uq.edu.au)