SUMMARY
Preprint: bioRxiv
SiRCle (Signature Regulatory Clustering) model integration reveals mechanisms of phenotype regulation in renal cancer
Ariane Mora^1, Christina Schmidt^2,3, Brad Balderson1, Christian Frezza3#, Mikael Bodén1#
School of Chemistry and Molecular Biosciences, University of Queensland, Molecular Biosciences Building 76, St Lucia QLD 4072, Australia.
Medical Research Council Cancer Unit, University of Cambridge, Hutchison/MRC Research Centre, Box 197, Cambridge Biomedical Campus, Cambridge CB2 0X2, United Kingdom
CECAD Research Center, University Hospital Cologne, Joseph-Stelzmann-Str. 26, 50931 Cologne, Germany
^Joint first authors #Joint last authors
Note Christina and Ariane are equal joint first authors and the authors may swap the order of their names as they so choose :)
Abstract
Clear cell renal cell carcinoma (ccRCC) tumours develop and progress via complex remodelling of the kidney epigenome, transcriptome, proteome, and metabolome. Given the subsequent tumour and inter-patient heterogeneity, drug-based treatments report limited success, calling for multi-omics studies to extract regulatory relationships, and ultimately, to develop targeted therapies. However, current methods are unable to extract nonlinear multi-omics perturbations.
Here, we present SiRCle (Signature Regulatory Clustering), a novel method to integrate DNA methylation, RNA-seq and proteomics data. Applying SiRCle to a case study of ccRCC, we disentangle the layer (DNA methylation, transcription and/or translation) where dysregulation first occurs and find the primary biological processes altered. Next, we detect regulatory differences between patient subsets by using a variational autoencoder to integrate omics’ data followed by statistical comparisons on the integrated space. In ccRCC patients, SiRCle allows to identify metabolic enzymes and cell-type-specific markers associated with survival along with the likely molecular driver behind the gene’s perturbations.
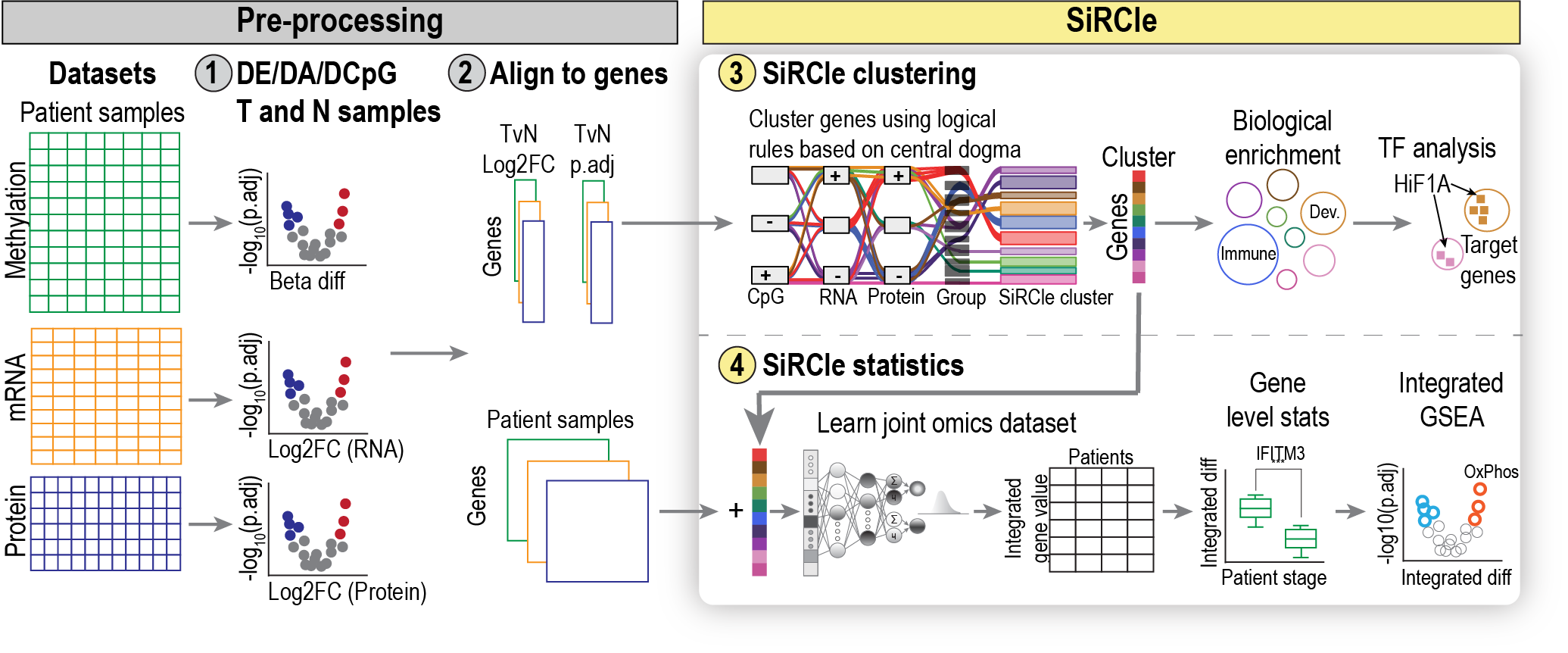
The general table of how we define regulatory clusters.
"""
| Methylation | RNAseq | Proteomics | Regulation driver_1 | Regulation driver_2 | Regulation_Grouping1 | Regulation_Grouping2 | Regulation_Grouping3 |
|------------------|-----------|------------|------------------------------|-------------------------|----------------------|----------------------|----------------------|
| Hypermethylation | DOWN | DOWN | Methylation increase (MDS) | None | MDS | MDS | MDS |
| Hypermethylation | UP | DOWN | mRNA increase (TPDE) | Protein decrease (TMDS) | TPDE+TMDS | TPDE+TMDS | TMDS |
| Hypermethylation | UP | UP | mRNA increase (TPDE) | None | TPDE | TPDE | TPDE |
| Hypermethylation | DOWN | UP | Methylation increase (MDS) | Protein increase (TMDE) | MDS+TMDE | TMDE | TMDE |
| Hypermethylation | No Change | UP | mRNA increase (TPDE) | Protein increase (TMDE) | TPDE+TMDE | TMDE | TMDE |
| Hypermethylation | No Change | DOWN | mRNA increase (TPDE) | Protein decrease (TMDS) | TPDE+TMDS | TMDS | TMDS |
| Hypermethylation | UP | No Change | mRNA increase (TPDE) | Protein decrease (TMDS) | TPDE+TMDS | TPDE+TMDS | TMDS |
| Hypermethylation | DOWN | No Change | Methylation increase (MDS) | Protein increase (TMDE) | MDS+TMDE | MDS+TMDE | TMDE |
| Hypermethylation | No Change | No Change | Methylation increase (ncRNA) | None | MDS-ncRNA | MDS_ncRNA | MDS_ncRNA |
| Hypomethylation | DOWN | DOWN | mRNA decrease (TPDS) | None | TPDS | TPDS | TPDS |
| Hypomethylation | UP | DOWN | Methylation decrease (MDE) | Protein decrease (TMDS) | MDE+TMDS | TMDS | TMDS |
| Hypomethylation | UP | UP | Methylation decrease (MDE) | None | MDE | MDE | MDE |
| Hypomethylation | DOWN | UP | mRNA decrease (TPDS) | Protein increase (TMDE) | TPDS+TMDE | TPDS+TMDE | TMDE |
| Hypomethylation | No Change | UP | mRNA decrease (TPDS) | Protein increase (TMDE) | TPDS+TMDE | TMDE | TMDE |
| Hypomethylation | No Change | DOWN | mRNA decrease (TPDS) | Protein decrease (TMDS) | TPDS+TMDS | TMDS | TMDS |
| Hypomethylation | UP | No Change | Methylation decrease (MDE) | Protein decrease (TMDS) | MDE+TMDS | MDE+TMDS | TMDS |
| Hypomethylation | DOWN | No Change | mRNA decrease (TPDS) | Protein increase (TMDE) | TPDS+TMDE | TPDS+TMDE | TMDE |
| Hypomethylation | No Change | No Change | Methylation decrease (ncRNA) | None | MDE+ncRNA | MDE_ncRNA | MDE_ncRNA |
| No Change | DOWN | UP | mRNA decrease (TPDS) | Protein increase (TMDE) | TPDS+TMDE | TPDS+TMDE | TMDE |
| No Change | UP | DOWN | mRNA increase (TPDE) | Protein decrease (TMDS) | TPDE+TMDS | TPDE+TMDS | TMDS |
| No Change | DOWN | DOWN | mRNA decrease (TPDS) | None | TPDS | TPDS | TPDS |
| No Change | UP | UP | mRNA increase (TPDE) | None | TPDE | TPDE | TPDE |
| No Change | No Change | UP | Protein increase (TMDE) | None | TMDE | TMDE | TMDE |
| No Change | No Change | DOWN | Protein decrease (TMDS) | None | TMDS | TMDS | TMDS |
| No Change | UP | No Change | mRNA increase (TPDE) | Protein decrease (TMDS) | TPDE+TMDS | TPDE+TMDS | TMDS |
| No Change | DOWN | No Change | mRNA decrease (TPDS) | Protein increase (TMDE) | TPDS+TMDE | TPDS+TMDE | TMDE |
| No Change | No Change | No Change | NoChange | NoChange | NoChange | NoChange | NoChange |
"""
Please post questions and issues related to sci-rcm on the Issues section of the GitHub repository.
Reproducibility
- Notebook RCM Part 1 ccRCC Figure 1
- Notebook RCM Part 1 ccRCC Figure 2
- Notebook RCM Part 1 ccRCC Figure 3
- Notebook RCM Part 1 ccRCC Figure 4
- Notebook RCM Part 1 ccRCC Figure 5
- Notebook RCM Part 1 ccRCC Figure 5b
- Notebook RCM Part 1 ccRCC S.Figure 1
- Notebook RCM Part 1 PanCan Figure 1
- Notebook RCM Part 1 PanCan Figure 2
- Notebook RCM Part 1 PanCan Figure 3
- Notebook RCM Part 1 PanCan Figure 4
- Notebook RCM Part 1 PanCan S.Figure 1
- Notebook VAE Part 2 ccRCC Figure 4
- Notebook RCM Part 1 PanCan Figure 5
- Notebook VAE Part 2 ccRCC Figure 6
- Notebook VAE Part 2 PanCan Figure 4
- Notebook VAE Part 2 PanCan Figure 5
- Notebook VAE Part 2 PanCan Figure 6
- Notebook Part 3 Comparison Figure 1
- Notebook Part 4 ITH Processing
- Notebook Part 4 ITH S.Figure 1
- Notebook Part 4 ITH Analysis
- Notebook Metabolism ccRCC
- Notebook for MOMIX benchmarking
- Notebook for RNA processing part 1
- Notebook for Clinical processing
- Protein Imputation
- Notebook for Protein processing
- Notebook for Methylation processing
- Notebook for Phospho-proteoimcs peptide processing
- Notebook for RNA processing part 2
- Notebook for Generating datasets
- Tumour vs normal comparison
- Notebook for Filtering CpGs to Genes
- Notebook for performing SiRCle clustering
- ORA for SiRCle clusters
- Notebook for Over Representation Analysis Visualisation for SiRCle
- Notebook for TF analysis
- Notebook for VAE integration
- GSEA on the integrated VAE value
- Set up single cell files
- Notebook for Single cell analysis using integrated genes Stage IV vs Stage I
- Notebook for Single cell analysis using integrated genes for PBRM1 vs BAP1
- Notebook for metabolomics analysis using publicly available data
- Notebook for RCM data with metabolic pathways
- Notebook for VAE data with metabolic pathways